Module for handling chemical reactions. More...
Functions/Subroutines | |
| subroutine, public | chemistry_initialize (tree, cfg) |
| Initialize module and load chemical reactions. | |
| subroutine, public | chemistry_convert_stochastic_species (tree, rng) |
| Convert stochastic species into regular species, using random numbers. | |
| subroutine, public | chemistry_write_summary (fname) |
| Write a summary of the reactions (TODO) and the ionization and attachment coefficients (if working at constant pressure) | |
| subroutine, public | chemistry_get_breakdown_field (field_td, min_growth_rate) |
| Get the breakdown field in Townsend. | |
| subroutine, public | get_rates (fields, rates, n_cells, energy_ev) |
| Compute reaction rates. | |
| subroutine, public | get_derivatives (dens, rates, derivs, n_cells) |
| Compute derivatives due to chemical reactions. Note that the 'rates' argument is modified. | |
| subroutine, public | read_reactions (filename, read_success) |
| Read reactions from a file. | |
| elemental integer function, public | species_index (name) |
| Find index of a species, return -1 if not found. | |
Variables | |
| integer, parameter, public | ionization_reaction = 1 |
| Identifier for ionization reactions. | |
| integer, parameter, public | attachment_reaction = 2 |
| Identifier for attachment reactions. | |
| integer, parameter, public | recombination_reaction = 3 |
| Identifier for recombination reactions. | |
| integer, parameter, public | detachment_reaction = 4 |
| Identifier for detachment reactions. | |
| integer, parameter, public | general_reaction = 5 |
| Identifier for general reactions (not of any particular type) | |
| character(len=20), dimension(*), parameter, public | reaction_names = [character(len=20) :: "ionization", "attachment", "recombination", "detachment", "general"] |
| integer, parameter | rate_tabulated_energy = 0 |
| Reaction with a field-dependent reaction rate. | |
| integer, public, protected | n_species = 0 |
| Number of species present. | |
| integer, public, protected | n_gas_species = 0 |
| Number of gas species present. | |
| integer, public, protected | n_plasma_species = 0 |
| Number of plasma species present. | |
| integer, public, protected | n_reactions = 0 |
| Number of reactions present. | |
| integer, public, protected | n_stochastic_species = 0 |
| Number of stochastic species. | |
| character(len=comp_len), dimension(max_num_species), public, protected | species_list |
| List of the species. | |
| integer, dimension(max_num_species), public, protected | species_charge = 0 |
| Charge of the species. | |
| integer, dimension(max_num_species), public, protected | species_itree |
| species_itree(n) holds the index of species n in the tree (cell-centered variables) | |
| type(reaction_t), dimension(max_num_reactions), public, protected | reactions |
| List of reactions. | |
| integer, dimension(:), allocatable, public, protected | charged_species_itree |
| List with indices of charged species. | |
| integer, dimension(:), allocatable, public, protected | charged_species_charge |
| List with charges of charged species. | |
Detailed Description
Module for handling chemical reactions.
Function/Subroutine Documentation
◆ chemistry_convert_stochastic_species()
| subroutine, public m_chemistry::chemistry_convert_stochastic_species | ( | type(af_t), intent(inout) | tree, |
| type(rng_t), intent(inout) | rng | ||
| ) |
Convert stochastic species into regular species, using random numbers.
- Parameters
-
[in,out] rng Random number generator
Definition at line 435 of file m_chemistry.f90.
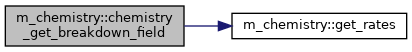
◆ chemistry_get_breakdown_field()
| subroutine, public m_chemistry::chemistry_get_breakdown_field | ( | real(dp), intent(out) | field_td, |
| real(dp), intent(in) | min_growth_rate | ||
| ) |
Get the breakdown field in Townsend.
- Parameters
-
[out] field_td Breakdown field in Townsend [in] min_growth_rate Minimal growth rate for breakdown
Definition at line 622 of file m_chemistry.f90.
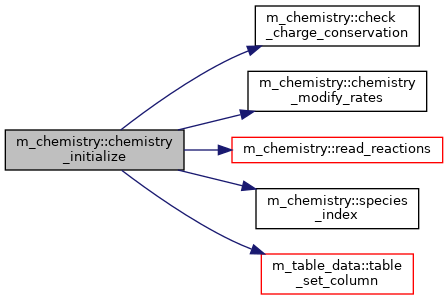
◆ chemistry_initialize()
| subroutine, public m_chemistry::chemistry_initialize | ( | type(af_t), intent(inout) | tree, |
| type(cfg_t), intent(inout) | cfg | ||
| ) |
Initialize module and load chemical reactions.
Definition at line 192 of file m_chemistry.f90.
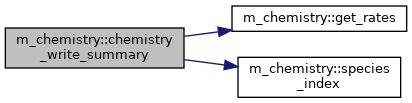
◆ chemistry_write_summary()
| subroutine, public m_chemistry::chemistry_write_summary | ( | character(len=*), intent(in) | fname | ) |
Write a summary of the reactions (TODO) and the ionization and attachment coefficients (if working at constant pressure)
Definition at line 532 of file m_chemistry.f90.
◆ get_derivatives()
| subroutine, public m_chemistry::get_derivatives | ( | real(dp), dimension(n_cells, n_species), intent(in) | dens, |
| real(dp), dimension(n_cells, n_reactions), intent(inout) | rates, | ||
| real(dp), dimension(n_cells, n_species), intent(out) | derivs, | ||
| integer, intent(in) | n_cells | ||
| ) |
Compute derivatives due to chemical reactions. Note that the 'rates' argument is modified.
- Parameters
-
[in] dens Species densities [in,out] rates On input, reaction rate coefficients. On output, actual reaction rates. [out] derivs Derivatives of the chemical species
Definition at line 761 of file m_chemistry.f90.
◆ get_rates()
| subroutine, public m_chemistry::get_rates | ( | real(dp), dimension(n_cells), intent(in) | fields, |
| real(dp), dimension(n_cells, n_reactions), intent(out) | rates, | ||
| integer, intent(in) | n_cells, | ||
| real(dp), dimension(n_cells), intent(in), optional | energy_ev | ||
| ) |
Compute reaction rates.
- Todo:
- These reactions do not take into account a variable gas_temperature
- Parameters
-
[in] n_cells Number of cells [in] fields The field (in Td) in the cells [out] rates The reaction rates [in] energy_ev Electron energy in eV
Definition at line 669 of file m_chemistry.f90.
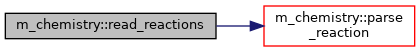
◆ read_reactions()
| subroutine, public m_chemistry::read_reactions | ( | character(len=*), intent(in) | filename, |
| logical, intent(out) | read_success | ||
| ) |
Read reactions from a file.
Definition at line 845 of file m_chemistry.f90.
◆ species_index()
| elemental integer function, public m_chemistry::species_index | ( | character(len=*), intent(in) | name | ) |
Find index of a species, return -1 if not found.
Definition at line 1265 of file m_chemistry.f90.

Variable Documentation
◆ attachment_reaction
| integer, parameter, public m_chemistry::attachment_reaction = 2 |
Identifier for attachment reactions.
Definition at line 18 of file m_chemistry.f90.
◆ charged_species_charge
| integer, dimension(:), allocatable, public, protected m_chemistry::charged_species_charge |
List with charges of charged species.
Definition at line 174 of file m_chemistry.f90.
◆ charged_species_itree
| integer, dimension(:), allocatable, public, protected m_chemistry::charged_species_itree |
List with indices of charged species.
Definition at line 171 of file m_chemistry.f90.
◆ detachment_reaction
| integer, parameter, public m_chemistry::detachment_reaction = 4 |
Identifier for detachment reactions.
Definition at line 22 of file m_chemistry.f90.
◆ general_reaction
| integer, parameter, public m_chemistry::general_reaction = 5 |
Identifier for general reactions (not of any particular type)
Definition at line 24 of file m_chemistry.f90.
◆ ionization_reaction
| integer, parameter, public m_chemistry::ionization_reaction = 1 |
Identifier for ionization reactions.
Definition at line 16 of file m_chemistry.f90.
◆ n_gas_species
| integer, public, protected m_chemistry::n_gas_species = 0 |
Number of gas species present.
Definition at line 135 of file m_chemistry.f90.
◆ n_plasma_species
| integer, public, protected m_chemistry::n_plasma_species = 0 |
Number of plasma species present.
Definition at line 138 of file m_chemistry.f90.
◆ n_reactions
| integer, public, protected m_chemistry::n_reactions = 0 |
Number of reactions present.
Definition at line 141 of file m_chemistry.f90.
◆ n_species
| integer, public, protected m_chemistry::n_species = 0 |
Number of species present.
Definition at line 132 of file m_chemistry.f90.
◆ n_stochastic_species
| integer, public, protected m_chemistry::n_stochastic_species = 0 |
Number of stochastic species.
Definition at line 144 of file m_chemistry.f90.
◆ rate_tabulated_energy
| integer, parameter m_chemistry::rate_tabulated_energy = 0 |
Reaction with a field-dependent reaction rate.
Definition at line 66 of file m_chemistry.f90.
◆ reaction_names
| character(len=20), dimension(*), parameter, public m_chemistry::reaction_names = [character(len=20) :: "ionization", "attachment", "recombination", "detachment", "general"] |
Definition at line 26 of file m_chemistry.f90.
◆ reactions
| type(reaction_t), dimension(max_num_reactions), public, protected m_chemistry::reactions |
List of reactions.
Definition at line 159 of file m_chemistry.f90.
◆ recombination_reaction
| integer, parameter, public m_chemistry::recombination_reaction = 3 |
Identifier for recombination reactions.
Definition at line 20 of file m_chemistry.f90.
◆ species_charge
| integer, dimension(max_num_species), public, protected m_chemistry::species_charge = 0 |
Charge of the species.
Definition at line 153 of file m_chemistry.f90.
◆ species_itree
| integer, dimension(max_num_species), public, protected m_chemistry::species_itree |
species_itree(n) holds the index of species n in the tree (cell-centered variables)
Definition at line 156 of file m_chemistry.f90.
◆ species_list
| character(len=comp_len), dimension(max_num_species), public, protected m_chemistry::species_list |
List of the species.
Definition at line 147 of file m_chemistry.f90.